UmbrellaSampling
Purpose
Umbrella sampling refers to a group of simulation methods designed to enhance configurational sampling so that the user can accurately calculate thermodynamic metrics of the molecular system that they are working with. Calculating accurate thermodynamics requires that the behavior of the system be reproduced multiple times so that all of the possible dynamics are captured. For example, if the goal of a project is to calculate the binding behavior between two proteins, it is necessary to simulate the binding and unbinding events many times so that essentially every conceivable way the two proteins interact with each other is captured at some point. The system could theoretically be simulated without any special methods for a very long time to replicate the binding/unbinding process. However, this is impractical because the amount of simulation time required to allow two free proteins to bind and unbind would be enormous. Moreover, unbinding events may be impossible to capture if the interaction turns out to be particularly strong.
Umbrella simulation methods speed up the sampling process by forcing the system to sample dynamics even at unfavorable configurations, such as a configuration that results in a local maxima in the system energy. They do this by biasing the system to stay near a desired setup, typically through some sort of restraint.
Typical Setup
Often, in Dr. Knotts' group, umbrella sampling refers to using a harmonic restraint to help hold two atoms (or two molecules) at a specified separation distance. A series of independent simulations are set up where the harmonic restraint distance is changed incrementally from short distances to long distances over the course of the simulation series. Each simulated restraint distance is independent of the others around it, so that the entire series can be run in parallel at one time. Each simulation produces a histogram of the actual restraint values experienced during the simulation run, and these outputs are combined together to produce the free energy of the system as a function of the restraint distances. The figure below helps to illustrate the connection between the simulations and the final free energy plot.
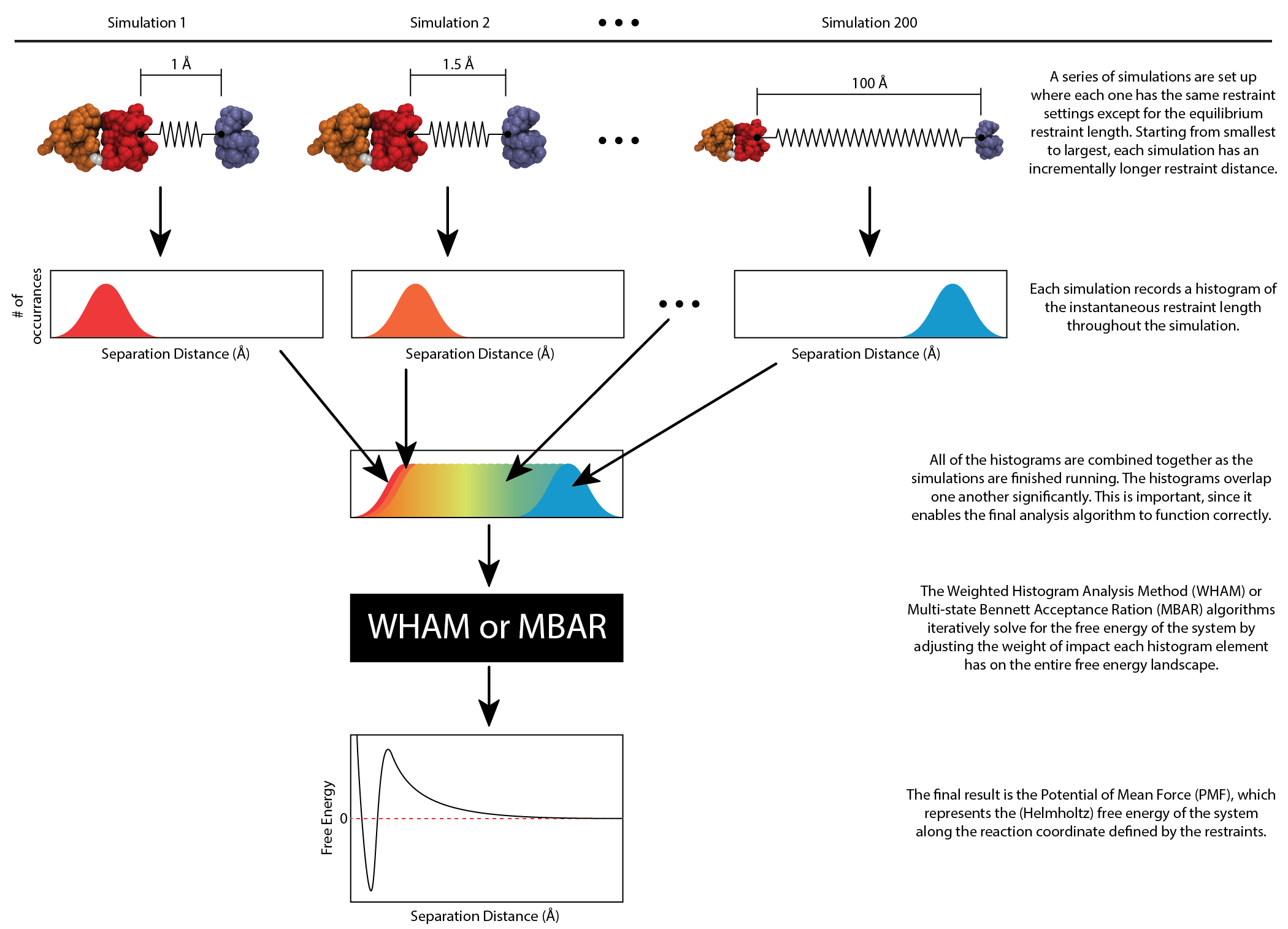
Flowchart showing the relationship between individual umbrella sampling simulations and the resulting system free energy.
Setup
Number of Required Simulations
Umbrella sampling typically requires a large number of simulations. The density of the simulations also increases as the umbrella series goes from 1D to 2D to 3D. For example, for a 1D umbrella where the reaction coordinate starts at 10 Å and ends at 100 Å, the harmonic restraint equilibrium lengths will likely need to be 10.0 Å, 12.5 Å, 15.0 Å, ..., 100 Å. If a second reaction coordinate is added, the spacing between umbrellas should decrease to avoid striations from appearing in the resulting 2D PMF. For example, the new equilibrium lengths would be something like 10.0 Å, 11.5 Å, 13.0 Å, ..., 100.0 Å for both reaction coordinates.
Harmonic Spring Constants
The strength of the harmonic restraint is an important factor when setting up umbrella simulations. There is no way to mathematically determine a priori what the spring constant should be, since there are several factors involved. For example, if a strong spring constant (e.g. 5 kcal mol-1) is used for each harmonic restraint, umbrellas will need to be spaced very close together. In contrast, if a weaker spring constant is used (e.g. 0.5 kcal mol-1), then the umbrellas can be spaced further apart. The ultimate goal is to ensure that, for the total number of time steps, that the histograms from each umbrella adequately overlaps neighboring histograms. As such, the user should perform a few initial simulations and check the histograms to ensure that the spacing between the umbrellas works well with the chosen spring constant.
In general, strong spring constants help ensure that very unfavorable configurations are sampled properly, but they require that many more simulations be performed to ensure proper histogram overlap. Weaker spring constants mean that fewer computer resources are needed, but it may be difficult to totally sample very unfavorable states.
In reality, the user can mix and match restraint equilibrium lengths and spring constants. For example, in phase space regions that the user knowns involve unfavorable configurations, more umbrellas with stronger spring constants can be used. In areas where molecular configurations are neither favorable nor unfavorable (such as long separation distances between molecules) that are neutral can involve fewer umbrellas with weaker spring constants. Note that a mixed setup as just described, while reducing the computational resources that are needed, complicates the post processing tasks. The remainder of this page, as well as example scripts, assume that every umbrella uses the same spring constant value.
Automating the Setup
As demonstrated above, umbrella sampling requires that the user set up a large number of simulations with unique settings. This task can be easily automated using BASH scripts. It is likely that the only real unique